Newly Discovered RNA Dysregulation Process Contributes to Neurodegeneration, Study Finds
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
- fact-checked
- peer-reviewed publication
- trusted source
- proofread
by Ingrid Fadelli , Medical Xpress
Recent neuroscience studies have consistently outlined the role of the C9ORF72 gene in the development of some neurodegenerative diseases. These studies found that mutations of this gene increase the risk of developing amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), two neurodegenerative disorders characterized by motor impairments, issues with communicating and other distinct symptoms.
Researchers at Johns Hopkins University School of Medicine, University of Chicago, the Howard Hughes Medical Institute and other institutes in the United States recently carried out a study aimed at better understanding the processes through which C9ORF72 gene mutation might ultimately contribute to the development of ALS and FTD. Their findings, published in Nature Neuroscience, highlight a promising avenue that could improve therapeutic interventions for these complex neurodegenerative disorders.
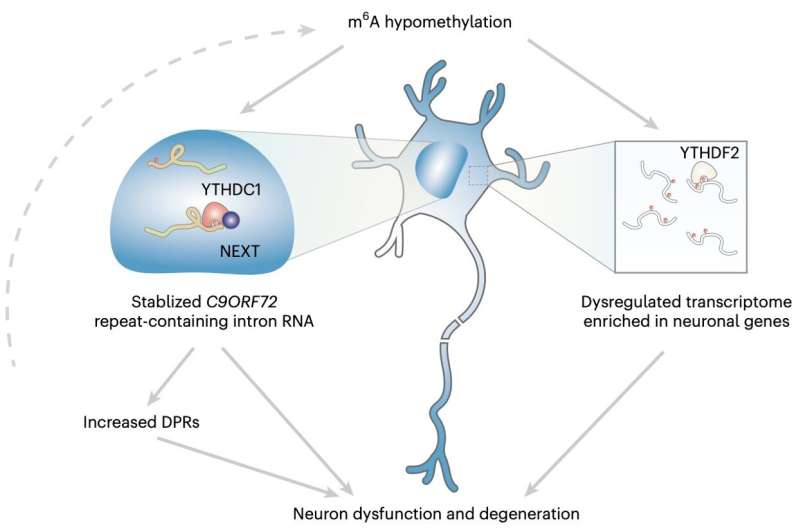
'My research group has been interested in identifying modifiers of the C9ORF72 repeat RNA metabolism,' Dr. Shuying Sun, one of the researchers who carried out the study, told Medical Xpress. 'A hexanucleotide repeat expansion in the C9ORF72 gene is the most prevalent genetic cause of both ALS and FTD. As the expansion is located in the noncoding region, the repeat-containing RNA is believed to be one major factor that drives the disease pathogenesis.'
Sun and her colleagues have been trying to identify genetic modifiers of the unconventional production of toxic poly-dipeptide proteins from the repeat RNA. This could in turn allow them to identify viable strategies to prevent the accumulation of these toxic proteins and thus reduce their toxic effects. In one of their previous studies, they thus performed a genome-wide CRISPR-Cas9 screen to search for genetic modifiers of dipeptide production.
'We found that the two m6A methyltransferases are candidate genes that can modulate the poly-dipeptide level,' Sun explained. 'We also analyzed public proteomic data and found the two methyltransferases are downregulated in C9ORF72-ALS/FTD patient iPS-neurons. Altogether, this initiated our further study on the m6A dysregulation and how this contributes to neurodegeneration in C9ORF72-ALS/FTD.'
Sun and her colleagues performed a series of analyses on patient-derived induced pluripotent stem cell (iPSC)-differentiated neurons (iPSNs) and postmortem tissues extracted from deceased patients with ALS/FTD and with C9ORF72 gene mutations. Using various high-throughput genetic sequencing techniques, they specifically assessed an RNA modification known as M6A (methyladenosine), and its impact on the regulation of genes across the full range of expressed messenger RNA (mRNA), known as transcriptome.
'We also examined the molecular mechanism of m6A regulation on C9 repeat RNA,' Sun said. 'Finally, we measured neuron survival to examine the rescue efficacy by modulating the m6A pathway. Our findings reveal a novel layer of RNA regulation that plays a critical role in the pathogenic mechanism underlying neurodegeneration.'
Overall, the findings gathered by this research team showed that the m6A modification in the C9ORF72 genetic sequence of expanded repeats facilitated the decay of RNA via the nuclear reader YTHDC1, thus regulating the repeats. Moreover, reduction in m6A appeared to facilitate the accumulation of repeat RNAs and the encoding of toxic proteins known to be associated with neurodegeneration.
By elevating m6A methylation, Sun and her colleagues were able to significantly reduce the level of repeat RNAs, safeguarding the neurons they were experimenting on and reducing their risk of decaying. In the future, these results could pave the way towards the development of more effective treatments for ALS and FTD, for instance prompting trials assessing the value of epitranscriptomic therapy as a potential intervention.
'We would like to further test the rescue efficacy in vivo using appropriate mouse models,' Sun added. 'We are also interested in developing pharmaceutical approaches targeting this pathway. Additionally, we hope to investigate the underlying disease mechanisms involving RNA metabolism and m6A dysregulation in sporadic ALS and other neurological disorders.'
© 2023 Science X Network




