Exploring the Mechanism of Antioxidant Enzymes in Repairing DNA Damage
June 1, 2023
This article has been examined in accordance with Science X's editorial process and policies. Editors have ensured the credibility of the content by fact-checking and peer-reviewing the publication. They have also proofread the article. The Center for Genomic Regulation was responsible for this process.
A human cell is constantly active metabolically, carrying out chemical reactions that convert nutrients into energy and other useful products for the sustenance of life. Unfortunately, these reactions also produce reactive oxygen species which are dangerous by-products like hydrogen peroxide. Such species can damage the building blocks of DNA in a way that is comparable to the corrosion caused by oxygen and water on metal, resulting in rust. If DNA damage occurs, cells can pause momentarily and carry out repairs by synthesizing new building blocks and injecting them into the gaps. However, if enough damage accumulates, it can be catastrophic and threaten the genome's integrity much like the cumulative effect of rust causes buildings to collapse over time.
Cells are thought to contain their metabolic activity outside the nucleus within the cytoplasm and mitochondria to avoid damaging DNA. Additionally, antioxidant enzymes are deployed to mop up reactive oxygen species at their source before they reach DNA. This defensive strategy helps to protect the roughly 3 billion nucleotides from potentially catastrophic mutations. Despite the central role of cellular metabolism in maintaining genome integrity, there have been no systematic, unbiased studies on how metabolic perturbations affect the DNA damage and repair process. This gap in knowledge is particularly important in diseases like cancer, where metabolic processes are essential for unfettered growth.
Researchers led by Sara Sdelci at the Centre for Genomic Regulation (CRG) in Barcelona and Joanna Loizou at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences in Vienna have carried out various experiments to identify which metabolic enzymes and processes are essential for a cell's DNA damage response. The team's findings are published in the journal Molecular Systems Biology.
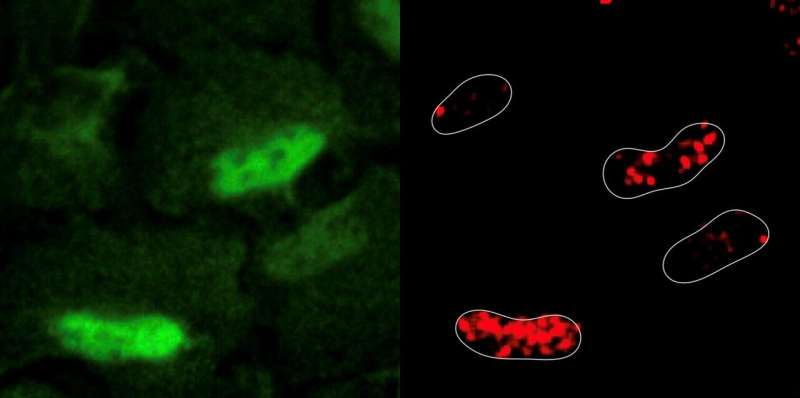
The researchers induced DNA damage in human cell lines using a common chemotherapy medication known as etoposide. Inducing DNA damage surprisingly resulted in reactive oxygen species accumulating inside the nucleus. The researchers observed cellular respiratory enzymes, which are a major source of reactive oxygen species, relocating from the mitochondria to the nucleus in response to DNA damage.
The researchers also used CRISPR-Cas9 to identify all the metabolic genes that were important for cell survival in this scenario. These experiments revealed that cells order the enzyme PRDX1, an antioxidant enzyme also normally found in mitochondria, to travel to the nucleus and scavenge the reactive oxygen species present to prevent further damage. PRDX1 was also found to repair the damage by regulating the cellular availability of aspartate, which is critical for synthesizing nucleotides, the building blocks of DNA.
Dr. Sdelci, corresponding author of the study and Group Leader at the Centre for Genomic Regulation, says, "'PRDX1 is like a robotic pool cleaner. Cells use it to keep their insides 'clean' and prevent the accumulation of reactive oxygen species, but never before at the nuclear level. This is evidence that in a state of crisis, the nucleus responds by appropriating mitochondrial machinery and establishing an emergency rapid-industrialization policy."
The findings represent a paradigm shift in cellular biology because it suggests that the nucleus may be metabolically active. The researchers state that the results can be useful in guiding future lines of cancer research, as some anti-cancer drugs, like etoposide, kill tumor cells by damaging their DNA and inhibiting the repair process. If enough damage accumulates, it initiates a process of autodestruction in the cancer cell.
During their experiments, the researchers found that knocking out metabolic genes critical for cellular respiration—the process that generates energy from oxygen and nutrients—made normal healthy cells become resistant to etoposide. The finding is important because many cancer cells are glycolytic, meaning that even in the presence of oxygen they generate energy without doing cellular respiration. This means etoposide, and other chemotherapies with a similar mechanism, is likely to have a limited effect in treating glycolytic tumors.
The authors of the study call for the exploration of new strategies such as dual treatment combining etoposide with drugs that also boost the generation of reactive oxygen species to overcome drug resistance and kill cancer cells faster. They also hypothesize that combining etoposide with inhibitors of nucleotide synthesis processes could potentiate the effect of the drug by preventing the repair of DNA damage and ensuring cancer cells self-destruct correctly.
Dr. Loizou, corresponding author and Group Leader at the Centre for Molecular Medicine and the Medical University of Vienna, highlights the value of taking data-driven approaches to uncover new biological processes. 'By using unbiased technologies such as CRISPR-Cas9 screening and metabolomics, we have learnt about how the two fundamental cellular processes of DNA repair and metabolism are intertwined. Our findings shed light on how targeting these two pathways in cancer might improve therapeutic outcomes for patients.'
Journal information: Molecular Systems Biology
Provided by Center for Genomic Regulation




