New Discoveries on Cat Evolution Unveiled by Genome Sequencing Project
November 2, 2023
The article received a thorough review, following the editorial procedure and guidelines of Science X, to ensure its credibility. The article has been:
- Fact-checked
- Identified as a peer-reviewed publication
- Verified as a trusted source
- Proofread
by Courtney Price from Texas A&M University.
The research was conducted at the Texas A&M School of Veterinary Medicine & Biomedical Sciences (VMBS) by an interdisciplinary team of researchers. They reveal new insights into the history of cat evolution, detailing how different species of cats, such as lions, tigers, and domestic cats, evolved and the connection between survival attributes in cats like the ability to smell prey and genetic changes.
With the help of genome comparation of several cat species, this research, now published in Nature Genetics, has enabled the researchers to understand why cat genomes tend to possess fewer complex changes in genetic makeup (like rearrangements of DNA parts) compared to other mammals like primates. They also discovered new insights about the parts of cat DNA likely to evolve quickly and their role in species differentiation.
"Our aim was to grasp the concept of how cats evolved and the genetic reasons for the differences in traits between different cat species," said Dr. Bill Murphy, a professor at VMBS who specializes in cat evolution. "We intended to leverage new technologies that allow us to map cat genomes more comprehensively."
He further added, "Our findings will support those who are studying feline diseases, behavior, and conservation by providing a thorough understanding of the genetic variations that distinguish each cat type."
The researchers focused their efforts to comprehend why feline chromosomes - cellular structures that carry genetic data for traits like size, fur color, sensory abilities - tend to remain stable compared to other mammals.
Explaining the findings, Murphy said, "For a while, it's been known that chromosomes of cats across various species are highly similar. For instance, chromosomes of domestic cats and lions are almost identical. There are fewer duplications, rearrangements, and other variations than we usually find in great apes."
In the primate order, such genetic variation has led to the evolution of different species like humans and great apes. The frequency of segmental duplications - the highly similar copies of segments of DNA found elsewhere in the genome - seems to be the key distinction between cats and apes.
"Our analysis of numerous cat genomes revealed that cats have far fewer segmental duplications compared to other mammals. In fact, primates have seven times more of these duplications than cats. This significant difference seems to explain why cat genomes are more stable," Murphy said.
Cats do have several variations in their DNA, despite having fewer major genetic rearrangements. Essential to their research, Murphy and team now comprehend which parts of cat DNA cause these variations, primarily the variations that cause the differences between species.
"There's a large region at the center of the X chromosome where the majority of genetic rearrangements occur. Moreover, it seems that a specific repetitive element within this region called DXZ4 is primarily responsible for the genetic isolation of at least two cat species - domestic and jungle cats," Murphy explained.
It seems that DXZ4, referred to by Murphy as a satellite repeat does not encode a physical trait like fur color. Instead, it aids in maintaining the three-dimensional structure of the X chromosome, playing a vital role in differentiating cat species.
'The exact mechanism is still unknown, but the comparison of countless cat genomes aids us in gauging the speed of DXZ4 evolution in one species in contrast with the others. The understanding we gained is that DXZ4 is among the most progressively evolving components of the cat genome, evolving at a pace faster than 99.5% of the rest of the genome,' he clarified.
'Due to its rapid mutation rate, we were successful in indicating why DXZ4 is presumably related to speciation,' stated Murphy.
The team, equipped with new, detailed genome sequences, found more explicit links between the quantity of olfactory genes, which regulate scent detection in cats, the variation in social behavior, and its correlation with their environment.
'As predators that heavily rely on their sense of smell to detect prey, it's pretty vital for cats, and we've always been keen to learn how genetic variation influences different cat species' ability to smell in their diverse environments,' said Murphy. 'The notable difference of odorant genes involved in detecting pheromones, signaling factors like identity, territory or danger, in lions and tigers is quite significant,' he added.
'Lions being highly sociable animals living in family groups and tigers leading a solitary life seems to contribute to this difference. The constant companionship of other lions could lean lions towards lesser dependence on pheromones and other odorants, as reflected by fewer genes of this kind in their genomes,' he explained.
On the contrary, tigers require a strong sense of smell to detect prey across large territories as well as locate mates.
'Overall, tigers possess enormous olfactory and pheromone receptor portfolios,' stated Murphy. 'We believe this is directly linked to the size of their territories and the diversity of their environments.'
However, domestic cats seem to have missed a broad range of olfactory genes.
'It would be logical for natural selection not to preserve these genes as domestic cats do not have to travel far to fulfill their needs due to human cohabitation,' he explained.
Murphy's favorite find from the study was the odorant receptors from fishing cats, an aquatic wild cat species residing in Southeast Asia.
'We showed that fishing cats have managed to keep many genes for detecting waterborne odorants, a trait seldom seen in terrestrial vertebrates,' he remarked. 'These specific genes have been lost with time in other cat species, but fishing cats still possess them.'
A new method of genome sequencing known as trio binning has enabled researchers to uncover this new information about olfactory genes in cats, by way of sequencing the most intricate regions of a genome.
This state-of-the-art technology also simplifies the process of separating maternal and paternal DNA.
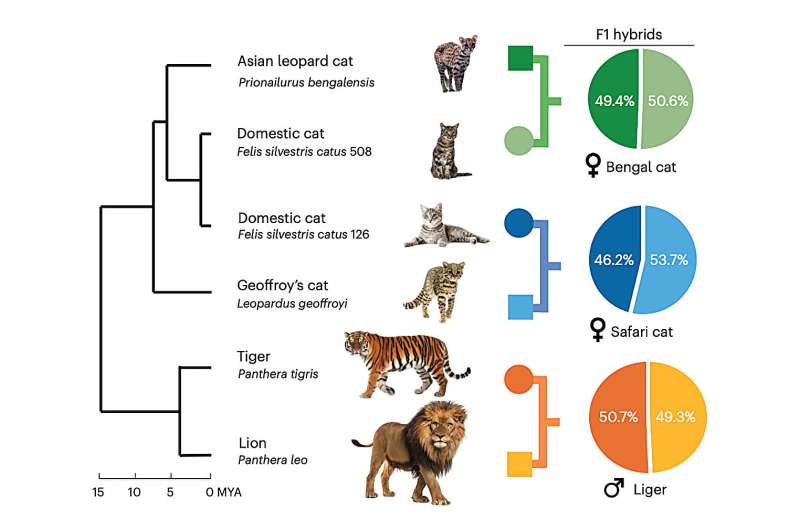
'Using trio binning, DNA from an F1 hybrid, an animal with DNA equally split between parents of different species, can be extracted and neatly divided into maternal and paternal DNA. This gives two thorough sets of DNA, one set for each parent species,' mentioned Murphy. 'This method is simpler and yields more comprehensive results.'
The most vital inference drawn from this study is that although many cat species may exhibit similarities, their differences are significant.
'These differences exemplify how these animals are flawlessly suited for their natural habitats,' explained Murphy. 'They aren't substitutable, providing valuable information for those working in conservation and restoration of species in their natural habitats.'
'Assuming that tigers from Sumatra and Siberia are similar would be incorrect,' he highlighted. 'Their environments are drastically different, and the respective tiger populations have probably developed specialized genetic adaptations to aid their survival in these diverse places.'
The fact that the sections of genomes most challenging to assemble may hold the key to understanding vital bodily systems like immunity and reproduction is equally significant for the scientific community.
'Immune and reproductive genes, along with olfactory genes, have been problematic to sequence and study. Past research lacked this kind of information. Consider how challenging it would be to research a genetic condition in cats, humans, or any species for that matter, without having access to all the elements; hence the significance of assembling complete genomes,' emphasized Murphy.
For now, Murphy and his team will persist in applying state-of-the-art genome sequencing and assembly technologies to cat genomes, with the aim of maximizing the understanding of the captivating world of cats.
The study was conceptualized by Bill Murphy—VMBS professor of veterinary integrative biosciences at Texas A&M and Wes Warren—professor of genomics in the Bond Life Sciences Center at the University of Missouri. Additional collaborations involved researchers from the University of Washington, University College Dublin, the Institute for Systems Biology in Seattle, Louisiana State University and the Guy Harvey Oceanographic Center.
Journal information: Nature Genetics
Provided by Texas A&M University




