Potential breakthrough in cell programming could revolutionize medical treatments
February 15, 2024
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
- fact-checked
- trusted source
- proofread
by Renata Solan, University of Wisconsin-Madison
Researchers can engineer cells to express new genes and produce specific proteins, giving the cells new parts to work with. But, it's much harder to provide cells with instructions on how to organize and use those new parts. Now, new tools from University of Wisconsin–Madison researchers offer an innovative way around this problem.
Their research is published in the journal Cell.
Everything a cell does depends on how molecules are organized within the cell. Inside our cells—all cells—proteins and other molecules undergo organization and reorganization to carry out cellular function. Like a fleet of commuter trains moving at scheduled intervals along their different routes, proteins within a cell are organized in time and space to carry out complex but predictable functions.
While the need to organize molecules within a cell is universal among living organisms, the specific proteins and mechanisms responsible for this organization vary. In a system specific to bacterial cells, for example, the proteins MinD and MinE—known collectively as MinDE—interact with each other along the cell membrane to produce wave-like patterns, which aid in the movement of molecules within the cell.
When molecules fail to organize properly within a cell, it can have serious consequences, including cells dividing unevenly and improper communication within and among cells, both of which are associated with developmental disorders and diseases such as cancer.
In short, we know how to give cells some new parts, but it's much harder to provide the instructions on how to organize and use them.
The mechanisms by which molecules organize and interact with each other have been fine-tuned over millennia of evolution. When scientists engineer cells to produce new molecules, it is difficult to get cells to use those new molecules without unintentionally disrupting other natural cellular functions.
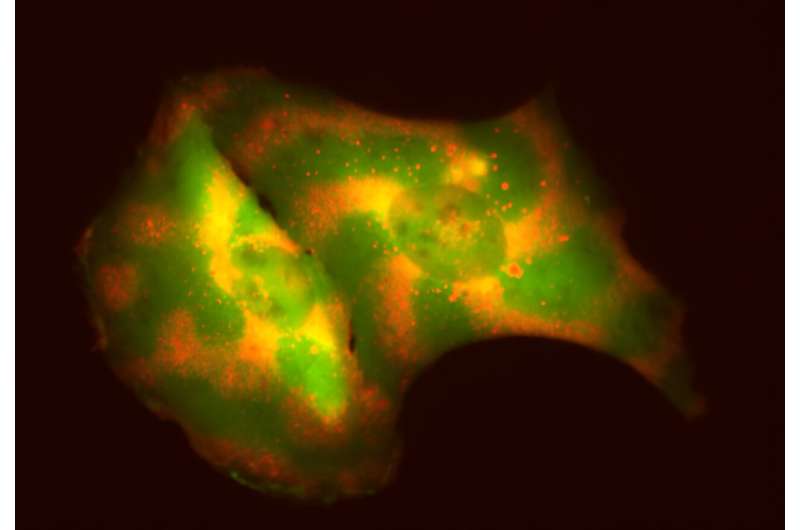
Biochemists at UW–Madison have developed a tool to control movement and organization of specific proteins in mammalian cells while leaving other proteins alone. Their new tool harnesses the waves and oscillations derived from interactions between MinDE proteins, which are found only in bacteria and do not interfere with mammalian cellular function.
By engineering interactions between the MinDE proteins and proteins of interest, the researchers created highly specified patterns to organize molecules within mammalian cells and induce cellular behaviors and functions. The tool allows researchers to tweak and alter the patterns in response to stimuli, essentially programming molecules to move around a cell to specific locations over time.
This innovative tool has multiple potential uses for scientists interested in engineering specific cellular activities or studying cellular activity in a living organism.
Controlling the ratio of MinDE proteins allows the researchers to design patterns of movement that determine how molecules are organized in a cell, which could be used to orchestrate cellular activities such as moving or communicating with other cells.
The variation in movement patterns can also be used to study cellular activity. As each ratio of MinDE proteins emits a unique oscillatory pattern, the proteins can be inserted into a group of cells to give each cell its own pattern—an individual beacon that lets researchers observe each cell more easily.
Scientists can also use the unique patterns to analyze the cell's signaling patterns in order to learn about the shape, location and signaling activity of each individual cell. UW–Madison's Coyle Lab researchers liken this use of the tool to an FM radio dial because it allows them to tune to the unique data that each cell emits in a multicellular system, a task that is usually very difficult.
The Coyle Lab plans to continue exploring the tool's applications, including studying the dynamics of signaling pathways in tumors, one classic example of cellular behavior and function gone wrong.
Provided by University of Wisconsin-Madison




