Starfish - Initially Assumed Headless Creatures - May Be Entirely Different
November 1, 2023
The article has undergone a thorough review process following Science X's editorial practices and norms. The content is confirmed to be credible, comprising highlighted features such as:
- fact-checked
- peer-reviewed publication
- proofread
by Chan Zuckerberg Biohub
Naturalists have been perplexed by the anatomy of a sea star or 'starfish' for ages trying to figure out its head or front end. The sea stars don't have an apparent head or tail because of their five identical arms. This distinctness in body pattern has led some to believe that sea stars don't have a head.
Stanford University and UC Berkeley labs, managed by Chan Zuckerberg Biohub San Francisco Investigators, recently released a study contradicting this belief. The study explains how the researchers detected gene signatures connected with head development in juvenile sea stars, whereas genes pertaining to an animal's torso and tail sections were almost non-existent.
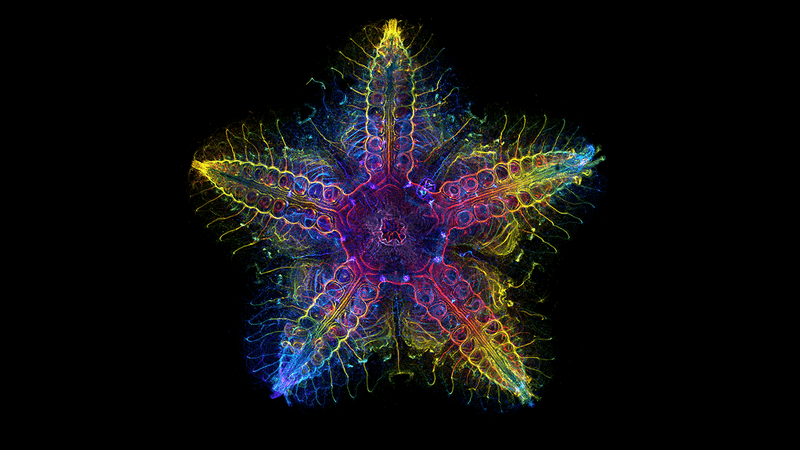
The research team employed advanced molecular and genomic methods to identify where different genes were expressed in the development and growth phases of sea stars. One team at Southampton utilized micro-CT scanning to scrutinize the animal's shape and structure meticulously.
In a surprising revelation, molecular signatures typically related to the foremost part of the head were found in the middle section of each sea star's arms. These signatures were found to be gradually more posterior moving towards the arms' edges.
The research, published in Nature on November 1, suggests that the sea stars evolved to only have heads overtime rather than not having a head.
Micro-CT scan displays the sea star's various parts – skeleton (gray), digestive system (yellow), nervous system (blue), muscles (red), and the water vascular system (purple). The credits are attributed to the University of Southampton.
According to Laurent Formery, the postdoctoral scholar and lead author of the study, a sea star is akin to a head without a trunk, moving along the seafloor. This finding significantly deviates from what scientists believed about these animals.
Christopher Lowe from Stanford University and Daniel Rokhsar from UC Berkeley, the study's marinologist, developmental biologist, and animal species evolution expert, have been collaborating for ten years.
The 1995 Nobel Prize in Physiology or Medicine was awarded to three scientists who proved that almost all animals, including humans, have bilateral symmetry. This body structure is believed to be formed by molecular switches, controlled by genes, expressed in specific head and trunk regions.
Most animal species, including vertebrates and many invertebrates, share this genetic program. However, the body structure of sea stars has often baffled scientists' comprehension of animal evolution. Sea stars display a five-fold symmetry with no conspicuous head or tail, unlike the bilateral symmetry shown by most animals. Determining the genetic programming that leads to this distinctive five-fold symmetry has remained an unresolved question.
Some scientists have speculated that the sea star's head-to-tail axis could extend from its back to its underbelly carpeted with tube feet. Others believe that each of the sea star's five arms signifies a cloned head-to-tail axis.
Efforts to conclusively validate these hypotheses have been confronted with challenges, mainly due to the incompatibility of gene expression detection methods used in a handful of model organisms. For years, Lowe and his team have endeavoured to utilize genetic information for mapping genetic activities across developing sea stars. But in the absence of comprehensive genetic tools developed through years of research, this process seemed considerably daunting.
Lowe encountered a solution for this problem at one of the regular San Francisco meetings of Biohub Investigators, where another researcher suggested he contact PacBio, a Silicon Valley–based company that builds genome-sequencing devices. Over the previous five years, PacBio had been perfecting a technique for sequencing massive quantities of genetic material using postage stamp–sized chips jam-packed with millions of individual chemical reactors, each primed to simultaneously read long stretches of DNA captured within.
Unlike traditional sequencing, which requires chopping genetic material into small pieces to ensure accuracy, PacBio's approach, called HiFi sequencing, can pull highly accurate data from intact, gene-sized DNA strands, making the process much faster and cheaper. It was exactly what Lowe and his team needed to establish a process for studying sea star genetics from the ground up.
'The kind of sequencing that would have taken months can now be done in a matter of hours, and it's hundreds of times cheaper than just five years ago,' said David Rank, also a co–senior author of the new study and a former PacBio Scientific Fellow. 'These advances meant we could start essentially from scratch in an organism that's not typically studied in the lab and put together the kind of detailed study that would have been impossible 10 years ago.'
This technology allowed the researchers to sequence the genomes of the sea stars and employ an approach called spatial transcriptomics, through which they could pinpoint which sea star genes are active at precise locations in the organism. To search for patterns that would indicate a head-to-tail axis, the researchers examined gene expression differences in three different directions across the body: from the sea star's center to its arm tips, from its top to its underbelly, and from one side edge of its arms to the other.
Then, to get a closer look at how certain key genes were behaving, they labeled them one by one with fluorescent dyes to create a detailed map of their distribution in the sea star body.
The researchers found that neither of the prominent hypotheses of sea star body plan structure was correct. Instead, they saw that gene expression corresponding to the forebrain in humans and other bilaterally symmetrical animals was located along the midline of sea stars' arms, with genetic expression corresponding to that of the human midbrain towards the arms' outer edges.
While the genes marking different subregions of the head in humans and other bilaterians were expressed in the sea star, only one of the genes typically associated with the trunk in animals was expressed, at the very edges of the sea stars' arms.
'These results suggest that the echinoderms, and sea stars in particular, have the most dramatic example of decoupling of the head and the trunk regions that we are aware of today,' said Formery, adding that some bizarre-looking sea star ancestors preserved in the fossil record do appear to have had a trunk. 'It just opens a ton of new questions that we can now start to explore.'
Questions that the team hopes to address next involve whether the genetic patterning seen in sea stars also shows up in sea urchins and sea cucumbers. For his part, Formery also wants to look into what the sea star can teach us about the evolution of the nervous system, which, he said, no one quite understands in echinoderms.
Learning more about the sea star and its relatives will not only help solve key mysteries of animal evolution, but could also inspire innovations in medicine, the researchers said. Sea stars walk by moving water through thousands of tube feet and digest their prey by extruding their stomachs outside of their bodies. It only stands to reason that these unusual creatures have also evolved completely unexpected strategies for staying healthy—which, if we took the time to understand them, could expand our approaches to combating human disease.
'It's certainly harder to work in organisms that are less frequently studied,' Rokhsar said. 'But if we take the opportunity to explore unusual animals that are operating in unusual ways, that means we are broadening our perspective of biology, which is eventually going to help us solve both ecological and biomedical problems.'
Journal information: Nature
Provided by Chan Zuckerberg Biohub




